在以前整理的GROMACS分子动力学模拟教程: 多肽-蛋白相互作用中, 提到了一种使用gmx sham计算自由能形貌图的简单方法. 这种方法适用于计算那种和两个变量相关的自由能形貌图, 并且容易作图. 有些文章中给出的就是这种图, 比如很多蛋白的自由能形貌图中给出了自由能与回旋半径以及RMSD的关系, 并以彩色填充图的形式给出, 看起来很高端的样子. 实际上做这种图还是比较简单的, 下面我就给个示例.
模拟
我们使用一个28肽的小蛋白, BBA折叠多肽1fme来做示例:
<figure><script>var Mol1=new ChemDoodle.TransformCanvas3D('Mol-1',642,396);Mol1.specs.shapes_color = '#fff';Mol1.specs.backgroundColor = 'black';Mol1.specs.compass_display = true;Mol1.specs.set3DRepresentation('Ball and Stick');Mol1.specs.projectionPerspective_3D = false;Mol1.specs.proteins_ribbonCartoonize = true;Mol1.handle = null;Mol1.timeout = 15;Mol1.specs.crystals_unitCellLineWidth = 1.5;Mol1.specs.nucleics_residueColor = 'rainbow';Mol1.specs.proteins_residueColor= 'rainbow';Mol1.startAnimation = ChemDoodle._AnimatorCanvas.prototype.startAnimation;Mol1.stopAnimation = ChemDoodle._AnimatorCanvas.prototype.stopAnimation;Mol1.isRunning = ChemDoodle._AnimatorCanvas.prototype.isRunning;Mol1.dblclick = ChemDoodle.RotatorCanvas.prototype.dblclick;Mol1.nextFrame = function(delta){var matrix = [];ChemDoodle.lib.mat4.identity(matrix);var change = delta*Math.PI/15000;ChemDoodle.lib.mat4.rotate(matrix,change,[1,0,0]);ChemDoodle.lib.mat4.rotate(matrix,change,[0,1,0]);ChemDoodle.lib.mat4.rotate(matrix,change,[0,0,1]);ChemDoodle.lib.mat4.multiply(this.rotationMatrix, matrix)};var Fmol='HEADER DE NOVO PROTEIN 16-AUG-00 1FME \nTITLE SOLUTION STRUCTURE OF FSD-EY, A NOVEL PEPTIDE ASSUMING A \nTITLE 2 BETA-BETA-ALPHA FOLD \nCOMPND MOL_ID: 1; \nCOMPND 2 MOLECULE: FSD-EY PEPTIDE; \nCOMPND 3 CHAIN: A; \nCOMPND 4 ENGINEERED: YES \nSOURCE MOL_ID: 1; \nSOURCE 2 SYNTHETIC: YES; \nSOURCE 3 OTHER_DETAILS: POINT MUTANT OF SEQUENCE GENERATED BY ORBIT \nSOURCE 4 DESIGN PROCESS \nKEYWDS BETA-BETA-ALPHA, ZINC FINGER, FSD-1, DESIGNED PROTEIN, DE \nKEYWDS 2 NOVO PROTEIN \nEXPDTA SOLUTION NMR \nNUMMDL 34 \nAUTHOR C.A.SARISKY,S.L.MAYO \nREVDAT 2 24-FEB-09 1FME 1 VERSN \nREVDAT 1 21-APR-01 1FME 0 \nJRNL AUTH C.A.SARISKY,S.L.MAYO \nJRNL TITL THE BETA-BETA-ALPHA FOLD: EXPLORATIONS IN SEQUENCE \nJRNL TITL 2 SPACE. \nJRNL REF J.MOL.BIOL. V. 307 1411 2001 \nJRNL REFN ISSN 0022-2836 \nJRNL PMID 11292351 \nJRNL DOI 10.1006/JMBI.2000.4345 \nREMARK 1 \nREMARK 2 \nREMARK 2 RESOLUTION. NOT APPLICABLE. \nREMARK 3 \nREMARK 3 REFINEMENT. \nREMARK 3 PROGRAM : X-PLOR 3.857 \nREMARK 3 AUTHORS : BRUNGER \nREMARK 3 \nREMARK 3 OTHER REFINEMENT REMARKS: NULL \nREMARK 4 \nREMARK 4 1FME COMPLIES WITH FORMAT V. 3.15, 01-DEC-08 \nREMARK 100 \nREMARK 100 THIS ENTRY HAS BEEN PROCESSED BY RCSB ON 25-AUG-00. \nREMARK 100 THE RCSB ID CODE IS RCSB011711. \nREMARK 210 \nREMARK 210 EXPERIMENTAL DETAILS \nREMARK 210 EXPERIMENT TYPE : NMR \nREMARK 210 TEMPERATURE (KELVIN) : 280 \nREMARK 210 PH : 5.0 \nREMARK 210 IONIC STRENGTH : NULL \nREMARK 210 PRESSURE : AMBIENT \nREMARK 210 SAMPLE CONTENTS : 2 MM PEPTIDE; 2 MM PEPTIDE \nREMARK 210 \nREMARK 210 NMR EXPERIMENTS CONDUCTED : 2D NOESY, DQF-COSY, TOCSY \nREMARK 210 SPECTROMETER FIELD STRENGTH : 600 MHZ \nREMARK 210 SPECTROMETER MODEL : UNITYPLUS \nREMARK 210 SPECTROMETER MANUFACTURER : VARIAN \nREMARK 210 \nREMARK 210 STRUCTURE DETERMINATION. \nREMARK 210 SOFTWARE USED : ANSIG, VNMR 6.1 \nREMARK 210 METHOD USED : X-PLOR: DISTANCE GEOMETRY, \nREMARK 210 REFINEMENT \nREMARK 210 \nREMARK 210 CONFORMERS, NUMBER CALCULATED : 49 \nREMARK 210 CONFORMERS, NUMBER SUBMITTED : 34 \nREMARK 210 CONFORMERS, SELECTION CRITERIA : STRUCTURES WITH THE LEAST \nREMARK 210 RESTRAINT VIOLATIONS \nREMARK 210 \nREMARK 210 BEST REPRESENTATIVE CONFORMER IN THIS ENSEMBLE : NULL \nREMARK 210 \nREMARK 210 REMARK: NULL \nREMARK 215 \nREMARK 215 NMR STUDY \nREMARK 215 THE COORDINATES IN THIS ENTRY WERE GENERATED FROM SOLUTION \nREMARK 215 NMR DATA. PROTEIN DATA BANK CONVENTIONS REQUIRE THAT \nREMARK 215 CRYST1 AND SCALE RECORDS BE INCLUDED, BUT THE VALUES ON \nREMARK 215 THESE RECORDS ARE MEANINGLESS. \nREMARK 500 \nREMARK 500 GEOMETRY AND STEREOCHEMISTRY \nREMARK 500 SUBTOPIC: TORSION ANGLES \nREMARK 500 \nREMARK 500 TORSION ANGLES OUTSIDE THE EXPECTED RAMACHANDRAN REGIONS: \nREMARK 500 (M=MODEL NUMBER; RES=RESIDUE NAME; C=CHAIN IDENTIFIER; \nREMARK 500 SSEQ=SEQUENCE NUMBER; I=INSERTION CODE). \nREMARK 500 \nREMARK 500 STANDARD TABLE: \nREMARK 500 FORMAT:(10X,I3,1X,A3,1X,A1,I4,A1,4X,F7.2,3X,F7.2) \nREMARK 500 \nREMARK 500 EXPECTED VALUES: GJ KLEYWEGT AND TA JONES (1996). PHI/PSI- \nREMARK 500 CHOLOGY: RAMACHANDRAN REVISITED. STRUCTURE 4, 1395 - 1400 \nREMARK 500 \nREMARK 500 M RES CSSEQI PSI PHI \nREMARK 500 1 TYR A 7 -68.79 -126.50 \nREMARK 500 1 LYS A 8 59.16 -118.77 \nREMARK 500 1 PHE A 21 -74.37 -68.42 \nREMARK 500 2 TYR A 3 153.35 -48.62 \nREMARK 500 2 THR A 4 31.46 -98.45 \nREMARK 500 2 TYR A 7 -68.65 -124.11 \nREMARK 500 2 LYS A 8 60.97 -115.62 \nREMARK 500 2 PHE A 21 -73.17 -64.51 \nREMARK 500 3 TYR A 3 154.82 -49.72 \nREMARK 500 3 TYR A 7 -64.92 -124.37 \nREMARK 500 3 PHE A 21 -73.89 -63.47 \nREMARK 500 4 TYR A 7 -77.44 -126.49 \nREMARK 500 4 LYS A 8 53.61 -109.76 \nREMARK 500 5 ASN A 14 121.44 -170.73 \nREMARK 500 5 PHE A 21 -74.56 -68.78 \nREMARK 500 6 PHE A 21 -75.78 -71.17 \nREMARK 500 8 TYR A 3 171.06 -50.29 \nREMARK 500 8 ASN A 14 130.87 -175.09 \nREMARK 500 8 GLU A 17 -70.07 -61.47 \nREMARK 500 9 THR A 4 54.14 -107.27 \nREMARK 500 9 ALA A 5 -162.36 -58.54 \nREMARK 500 9 LYS A 6 150.70 176.87 \nREMARK 500 9 TYR A 7 -44.05 -142.30 \nREMARK 500 9 PHE A 21 -73.79 -69.30 \nREMARK 500 10 GLN A 2 -164.24 -104.78 \nREMARK 500 10 TYR A 3 162.25 -47.30 \nREMARK 500 10 PHE A 21 -73.65 -64.74 \nREMARK 500 11 GLN A 2 -168.74 -108.92 \nREMARK 500 11 TYR A 3 162.88 -47.49 \nREMARK 500 11 PHE A 21 -74.66 -63.63 \nREMARK 500 12 THR A 4 42.82 -109.43 \nREMARK 500 12 PHE A 21 -73.10 -65.62 \nREMARK 500 13 TYR A 7 -63.24 -139.55 \nREMARK 500 14 TYR A 3 179.12 -54.52 \nREMARK 500 14 TYR A 7 -77.50 -125.41 \nREMARK 500 14 LYS A 8 54.48 -118.15 \nREMARK 500 14 PHE A 25 -96.24 -59.33 \nREMARK 500 14 LYS A 26 -70.10 -151.86 \nREMARK 500 15 TYR A 3 176.71 -51.74 \nREMARK 500 15 TYR A 7 -75.71 -125.79 \nREMARK 500 15 LYS A 8 55.95 -118.54 \nREMARK 500 15 PHE A 25 -95.04 -55.15 \nREMARK 500 15 LYS A 26 -62.25 -151.45 \nREMARK 500 16 TYR A 7 -54.50 -124.39 \nREMARK 500 16 PHE A 25 -93.56 -50.13 \nREMARK 500 16 LYS A 26 -65.71 -147.39 \nREMARK 500 17 TYR A 3 165.34 -49.53 \nREMARK 500 17 PHE A 21 -73.90 -69.56 \nREMARK 500 18 TYR A 3 -170.39 -69.44 \nREMARK 500 18 THR A 4 -71.39 -108.25 \nREMARK 500 18 ALA A 5 111.22 48.54 \nREMARK 500 18 TYR A 7 -79.24 -123.99 \nREMARK 500 18 LYS A 8 59.41 -113.07 \nREMARK 500 19 THR A 4 -77.67 -110.20 \nREMARK 500 19 ALA A 5 110.43 54.79 \nREMARK 500 19 ASN A 14 139.67 -170.77 \nREMARK 500 19 PHE A 21 -76.11 -66.02 \nREMARK 500 20 THR A 4 -75.10 -111.24 \nREMARK 500 20 ALA A 5 117.11 55.70 \nREMARK 500 20 ASN A 14 133.33 -175.45 \nREMARK 500 20 PHE A 21 -74.25 -67.02 \nREMARK 500 21 THR A 4 57.90 -108.52 \nREMARK 500 21 ALA A 5 -162.41 -59.06 \nREMARK 500 21 LYS A 6 142.76 176.04 \nREMARK 500 21 LYS A 8 -62.77 68.21 \nREMARK 500 21 PHE A 21 -73.93 -68.92 \nREMARK 500 21 PHE A 25 67.64 -111.10 \nREMARK 500 22 TYR A 3 160.13 -47.71 \nREMARK 500 22 PHE A 21 -74.24 -80.86 \nREMARK 500 22 PHE A 25 -92.15 -59.57 \nREMARK 500 22 LYS A 26 -63.85 -153.43 \nREMARK 500 23 GLN A 2 -156.06 -95.69 \nREMARK 500 23 TYR A 3 144.21 -28.40 \nREMARK 500 23 THR A 4 27.52 -158.19 \nREMARK 500 23 LYS A 8 57.95 -154.03 \nREMARK 500 23 PHE A 21 -74.17 -58.78 \nREMARK 500 24 PHE A 21 -73.64 -67.87 \nREMARK 500 24 PHE A 25 -91.95 -56.22 \nREMARK 500 24 LYS A 26 -62.34 -147.63 \nREMARK 500 25 GLN A 2 -156.19 -163.37 \nREMARK 500 25 TYR A 3 141.39 -25.25 \nREMARK 500 25 THR A 4 25.23 -156.18 \nREMARK 500 25 TYR A 7 -60.13 -126.70 \nREMARK 500 26 TYR A 3 171.80 -54.99 \nREMARK 500 26 ASN A 14 137.84 -171.23 \nREMARK 500 26 PHE A 25 -94.84 -49.00 \nREMARK 500 26 LYS A 26 -67.64 -152.13 \nREMARK 500 27 GLN A 2 -154.84 -158.76 \nREMARK 500 27 TYR A 3 137.47 -30.02 \nREMARK 500 27 THR A 4 25.71 -158.26 \nREMARK 500 27 LYS A 8 57.67 -154.21 \nREMARK 500 27 ARG A 13 30.52 -98.35 \nREMARK 500 27 PHE A 21 -73.44 -61.23 \nREMARK 500 27 PHE A 25 65.12 -102.85 \nREMARK 500 28 GLN A 2 -156.39 -161.05 \nREMARK 500 28 TYR A 3 137.87 -22.67 \nREMARK 500 28 THR A 4 24.11 -156.47 \nREMARK 500 28 TYR A 7 -68.03 -125.72 \nREMARK 500 28 LYS A 8 51.44 -108.08 \nREMARK 500 28 PHE A 21 -72.78 -60.26 \nREMARK 500 28 PHE A 25 47.62 -91.79 \nREMARK 500 29 GLN A 2 -157.44 -137.95 \nREMARK 500 29 TYR A 3 145.64 -27.26 \nREMARK 500 29 THR A 4 21.88 -155.81 \nREMARK 500 29 PHE A 21 -74.73 -72.18 \nREMARK 500 30 GLN A 2 -158.25 53.75 \nREMARK 500 30 TYR A 3 136.36 -15.23 \nREMARK 500 30 THR A 4 21.65 -153.65 \nREMARK 500 30 PHE A 21 -75.02 -62.59 \nREMARK 500 31 GLN A 2 174.12 55.23 \nREMARK 500 31 TYR A 3 175.57 -52.30 \nREMARK 500 31 PHE A 25 -98.38 -54.54 \nREMARK 500 31 LYS A 26 -46.86 -153.56 \nREMARK 500 32 GLN A 2 -157.81 -140.11 \nREMARK 500 32 TYR A 3 140.22 -21.01 \nREMARK 500 32 THR A 4 21.45 -155.27 \nREMARK 500 32 ALA A 5 -161.48 -62.34 \nREMARK 500 32 LYS A 6 -27.66 -177.55 \nREMARK 500 32 TYR A 7 -89.55 54.16 \nREMARK 500 32 LYS A 8 -60.46 -94.16 \nREMARK 500 33 TYR A 3 -155.39 30.54 \nREMARK 500 33 THR A 4 33.67 93.92 \nREMARK 500 33 TYR A 7 -67.93 -124.44 \nREMARK 500 34 TYR A 3 -155.48 29.90 \nREMARK 500 34 THR A 4 35.74 89.03 \nREMARK 500 34 PHE A 21 -73.10 -64.52 \nREMARK 500 \nREMARK 500 REMARK: NULL \nREMARK 500 \nREMARK 500 GEOMETRY AND STEREOCHEMISTRY \nREMARK 500 SUBTOPIC: PLANAR GROUPS \nREMARK 500 \nREMARK 500 PLANAR GROUPS IN THE FOLLOWING RESIDUES HAVE A TOTAL \nREMARK 500 RMS DISTANCE OF ALL ATOMS FROM THE BEST-FIT PLANE \nREMARK 500 BY MORE THAN AN EXPECTED VALUE OF 6*RMSD, WITH AN \nREMARK 500 RMSD 0.02 ANGSTROMS, OR AT LEAST ONE ATOM HAS \nREMARK 500 AN RMSD GREATER THAN THIS VALUE \nREMARK 500 (M=MODEL NUMBER; RES=RESIDUE NAME; C=CHAIN IDENTIFIER; \nREMARK 500 SSEQ=SEQUENCE NUMBER; I=INSERTION CODE). \nREMARK 500 \nREMARK 500 M RES CSSEQI RMS TYPE \nREMARK 500 1 ARG A 10 0.29 SIDE_CHAIN \nREMARK 500 1 ARG A 13 0.32 SIDE_CHAIN \nREMARK 500 1 ARG A 19 0.21 SIDE_CHAIN \nREMARK 500 1 ARG A 28 0.32 SIDE_CHAIN \nREMARK 500 2 ARG A 10 0.26 SIDE_CHAIN \nREMARK 500 2 ARG A 13 0.25 SIDE_CHAIN \nREMARK 500 2 ARG A 19 0.29 SIDE_CHAIN \nREMARK 500 2 ARG A 28 0.21 SIDE_CHAIN \nREMARK 500 3 ARG A 10 0.28 SIDE_CHAIN \nREMARK 500 3 ARG A 13 0.30 SIDE_CHAIN \nREMARK 500 3 ARG A 19 0.29 SIDE_CHAIN \nREMARK 500 3 ARG A 28 0.21 SIDE_CHAIN \nREMARK 500 4 ARG A 10 0.22 SIDE_CHAIN \nREMARK 500 4 ARG A 13 0.24 SIDE_CHAIN \nREMARK 500 4 ARG A 19 0.20 SIDE_CHAIN \nREMARK 500 4 ARG A 28 0.29 SIDE_CHAIN \nREMARK 500 5 ARG A 10 0.31 SIDE_CHAIN \nREMARK 500 5 ARG A 13 0.23 SIDE_CHAIN \nREMARK 500 5 ARG A 19 0.31 SIDE_CHAIN \nREMARK 500 5 ARG A 28 0.31 SIDE_CHAIN \nREMARK 500 6 ARG A 10 0.28 SIDE_CHAIN \nREMARK 500 6 ARG A 13 0.21 SIDE_CHAIN \nREMARK 500 6 ARG A 19 0.23 SIDE_CHAIN \nREMARK 500 6 ARG A 28 0.23 SIDE_CHAIN \nREMARK 500 7 ARG A 10 0.28 SIDE_CHAIN \nREMARK 500 7 ARG A 13 0.26 SIDE_CHAIN \nREMARK 500 7 ARG A 19 0.28 SIDE_CHAIN \nREMARK 500 7 ARG A 28 0.32 SIDE_CHAIN \nREMARK 500 8 ARG A 10 0.22 SIDE_CHAIN \nREMARK 500 8 ARG A 13 0.30 SIDE_CHAIN \nREMARK 500 8 ARG A 19 0.29 SIDE_CHAIN \nREMARK 500 8 ARG A 28 0.28 SIDE_CHAIN \nREMARK 500 9 ARG A 10 0.21 SIDE_CHAIN \nREMARK 500 9 ARG A 13 0.29 SIDE_CHAIN \nREMARK 500 9 ARG A 19 0.32 SIDE_CHAIN \nREMARK 500 9 ARG A 28 0.28 SIDE_CHAIN \nREMARK 500 10 ARG A 10 0.29 SIDE_CHAIN \nREMARK 500 10 ARG A 13 0.31 SIDE_CHAIN \nREMARK 500 10 ARG A 19 0.23 SIDE_CHAIN \nREMARK 500 10 ARG A 28 0.31 SIDE_CHAIN \nREMARK 500 11 ARG A 10 0.32 SIDE_CHAIN \nREMARK 500 11 ARG A 13 0.27 SIDE_CHAIN \nREMARK 500 11 ARG A 19 0.21 SIDE_CHAIN \nREMARK 500 11 ARG A 28 0.32 SIDE_CHAIN \nREMARK 500 12 ARG A 10 0.30 SIDE_CHAIN \nREMARK 500 12 ARG A 13 0.32 SIDE_CHAIN \nREMARK 500 12 ARG A 19 0.30 SIDE_CHAIN \nREMARK 500 12 ARG A 28 0.32 SIDE_CHAIN \nREMARK 500 13 ARG A 10 0.21 SIDE_CHAIN \nREMARK 500 13 ARG A 13 0.27 SIDE_CHAIN \nREMARK 500 13 ARG A 19 0.23 SIDE_CHAIN \nREMARK 500 13 ARG A 28 0.22 SIDE_CHAIN \nREMARK 500 14 ARG A 10 0.27 SIDE_CHAIN \nREMARK 500 14 ARG A 13 0.23 SIDE_CHAIN \nREMARK 500 14 ARG A 19 0.22 SIDE_CHAIN \nREMARK 500 14 ARG A 28 0.32 SIDE_CHAIN \nREMARK 500 15 ARG A 10 0.28 SIDE_CHAIN \nREMARK 500 15 ARG A 13 0.25 SIDE_CHAIN \nREMARK 500 15 ARG A 19 0.32 SIDE_CHAIN \nREMARK 500 15 ARG A 28 0.29 SIDE_CHAIN \nREMARK 500 16 ARG A 10 0.21 SIDE_CHAIN \nREMARK 500 16 ARG A 13 0.27 SIDE_CHAIN \nREMARK 500 16 ARG A 19 0.32 SIDE_CHAIN \nREMARK 500 16 ARG A 28 0.22 SIDE_CHAIN \nREMARK 500 17 ARG A 10 0.23 SIDE_CHAIN \nREMARK 500 17 ARG A 13 0.23 SIDE_CHAIN \nREMARK 500 17 ARG A 19 0.28 SIDE_CHAIN \nREMARK 500 17 ARG A 28 0.26 SIDE_CHAIN \nREMARK 500 18 ARG A 10 0.21 SIDE_CHAIN \nREMARK 500 18 ARG A 13 0.21 SIDE_CHAIN \nREMARK 500 18 ARG A 19 0.31 SIDE_CHAIN \nREMARK 500 18 ARG A 28 0.21 SIDE_CHAIN \nREMARK 500 19 ARG A 10 0.32 SIDE_CHAIN \nREMARK 500 19 ARG A 13 0.24 SIDE_CHAIN \nREMARK 500 19 ARG A 19 0.32 SIDE_CHAIN \nREMARK 500 19 ARG A 28 0.30 SIDE_CHAIN \nREMARK 500 20 ARG A 10 0.30 SIDE_CHAIN \nREMARK 500 20 ARG A 13 0.32 SIDE_CHAIN \nREMARK 500 20 ARG A 19 0.20 SIDE_CHAIN \nREMARK 500 20 ARG A 28 0.23 SIDE_CHAIN \nREMARK 500 21 ARG A 10 0.31 SIDE_CHAIN \nREMARK 500 21 ARG A 13 0.23 SIDE_CHAIN \nREMARK 500 21 ARG A 19 0.30 SIDE_CHAIN \nREMARK 500 21 ARG A 28 0.24 SIDE_CHAIN \nREMARK 500 22 ARG A 10 0.32 SIDE_CHAIN \nREMARK 500 22 ARG A 13 0.27 SIDE_CHAIN \nREMARK 500 22 ARG A 19 0.31 SIDE_CHAIN \nREMARK 500 22 ARG A 28 0.29 SIDE_CHAIN \nREMARK 500 23 ARG A 10 0.29 SIDE_CHAIN \nREMARK 500 23 ARG A 13 0.32 SIDE_CHAIN \nREMARK 500 23 ARG A 19 0.28 SIDE_CHAIN \nREMARK 500 23 ARG A 28 0.27 SIDE_CHAIN \nREMARK 500 24 ARG A 10 0.25 SIDE_CHAIN \nREMARK 500 24 ARG A 13 0.23 SIDE_CHAIN \nREMARK 500 24 ARG A 19 0.27 SIDE_CHAIN \nREMARK 500 24 ARG A 28 0.32 SIDE_CHAIN \nREMARK 500 25 ARG A 10 0.28 SIDE_CHAIN \nREMARK 500 25 ARG A 13 0.30 SIDE_CHAIN \nREMARK 500 25 ARG A 19 0.32 SIDE_CHAIN \nREMARK 500 25 ARG A 28 0.30 SIDE_CHAIN \nREMARK 500 26 ARG A 10 0.23 SIDE_CHAIN \nREMARK 500 26 ARG A 13 0.25 SIDE_CHAIN \nREMARK 500 26 ARG A 19 0.28 SIDE_CHAIN \nREMARK 500 26 ARG A 28 0.31 SIDE_CHAIN \nREMARK 500 27 ARG A 10 0.24 SIDE_CHAIN \nREMARK 500 27 ARG A 13 0.29 SIDE_CHAIN \nREMARK 500 27 ARG A 19 0.32 SIDE_CHAIN \nREMARK 500 27 ARG A 28 0.28 SIDE_CHAIN \nREMARK 500 28 ARG A 10 0.32 SIDE_CHAIN \nREMARK 500 28 ARG A 13 0.32 SIDE_CHAIN \nREMARK 500 28 ARG A 19 0.25 SIDE_CHAIN \nREMARK 500 28 ARG A 28 0.26 SIDE_CHAIN \nREMARK 500 29 ARG A 10 0.27 SIDE_CHAIN \nREMARK 500 29 ARG A 13 0.30 SIDE_CHAIN \nREMARK 500 29 ARG A 19 0.28 SIDE_CHAIN \nREMARK 500 29 ARG A 28 0.23 SIDE_CHAIN \nREMARK 500 30 ARG A 10 0.32 SIDE_CHAIN \nREMARK 500 30 ARG A 13 0.23 SIDE_CHAIN \nREMARK 500 30 ARG A 19 0.31 SIDE_CHAIN \nREMARK 500 30 ARG A 28 0.27 SIDE_CHAIN \nREMARK 500 31 ARG A 10 0.32 SIDE_CHAIN \nREMARK 500 31 ARG A 13 0.27 SIDE_CHAIN \nREMARK 500 31 ARG A 19 0.29 SIDE_CHAIN \nREMARK 500 31 ARG A 28 0.25 SIDE_CHAIN \nREMARK 500 32 ARG A 10 0.26 SIDE_CHAIN \nREMARK 500 32 ARG A 13 0.22 SIDE_CHAIN \nREMARK 500 32 ARG A 19 0.31 SIDE_CHAIN \nREMARK 500 32 ARG A 28 0.30 SIDE_CHAIN \nREMARK 500 33 ARG A 10 0.32 SIDE_CHAIN \nREMARK 500 33 ARG A 13 0.27 SIDE_CHAIN \nREMARK 500 33 ARG A 19 0.25 SIDE_CHAIN \nREMARK 500 33 ARG A 28 0.26 SIDE_CHAIN \nREMARK 500 34 ARG A 10 0.29 SIDE_CHAIN \nREMARK 500 34 ARG A 13 0.30 SIDE_CHAIN \nREMARK 500 34 ARG A 19 0.31 SIDE_CHAIN \nREMARK 500 34 ARG A 28 0.28 SIDE_CHAIN \nREMARK 500 \nREMARK 500 REMARK: NULL \nDBREF 1FME A 1 28 PDB 1FME 1FME 1 28 \nSEQRES 1 A 28 GLU GLN TYR THR ALA LYS TYR LYS GLY ARG THR PHE ARG \nSEQRES 2 A 28 ASN GLU LYS GLU LEU ARG ASP PHE ILE GLU LYS PHE LYS \nSEQRES 3 A 28 GLY ARG \nHELIX 1 1 ASN A 14 PHE A 25 1 12 \nSHEET 1 A 2 ALA A 5 LYS A 6 0 \nSHEET 2 A 2 THR A 11 PHE A 12 -1 N PHE A 12 O ALA A 5 \nCRYST1 1.000 1.000 1.000 90.00 90.00 90.00 P 1 1 \nORIGX1 1.000000 0.000000 0.000000 0.00000 \nORIGX2 0.000000 1.000000 0.000000 0.00000 \nORIGX3 0.000000 0.000000 1.000000 0.00000 \nSCALE1 1.000000 0.000000 0.000000 0.00000 \nSCALE2 0.000000 1.000000 0.000000 0.00000 \nSCALE3 0.000000 0.000000 1.000000 0.00000 \nMODEL 1 \nATOM 1 N GLU A 1 -14.196 7.703 4.296 1.00 0.00 N \nATOM 2 CA GLU A 1 -13.829 6.504 3.488 1.00 0.00 C \nATOM 3 C GLU A 1 -12.882 6.904 2.352 1.00 0.00 C \nATOM 4 O GLU A 1 -13.313 7.259 1.271 1.00 0.00 O \nATOM 5 CB GLU A 1 -15.154 5.986 2.925 1.00 0.00 C \nATOM 6 CG GLU A 1 -15.153 4.456 2.939 1.00 0.00 C \nATOM 7 CD GLU A 1 -15.672 3.955 4.287 1.00 0.00 C \nATOM 8 OE1 GLU A 1 -15.276 4.514 5.297 1.00 0.00 O \nATOM 9 OE2 GLU A 1 -16.456 3.020 4.288 1.00 0.00 O \nATOM 10 H1 GLU A 1 -14.723 7.404 5.141 1.00 0.00 H \nATOM 11 H2 GLU A 1 -14.788 8.339 3.723 1.00 0.00 H \nATOM 12 H3 GLU A 1 -13.332 8.203 4.588 1.00 0.00 H \nATOM 13 HA GLU A 1 -13.374 5.753 4.113 1.00 0.00 H \nATOM 14 HB2 GLU A 1 -15.969 6.351 3.533 1.00 0.00 H \nATOM 15 HB3 GLU A 1 -15.276 6.336 1.911 1.00 0.00 H \nATOM 16 HG2 GLU A 1 -15.791 4.090 2.148 1.00 0.00 H \nATOM 17 HG3 GLU A 1 -14.146 4.095 2.786 1.00 0.00 H \nATOM 18 N GLN A 2 -11.596 6.846 2.593 1.00 0.00 N \nATOM 19 CA GLN A 2 -10.613 7.221 1.531 1.00 0.00 C \nATOM 20 C GLN A 2 -10.538 6.120 0.469 1.00 0.00 C \nATOM 21 O GLN A 2 -11.281 5.159 0.507 1.00 0.00 O \nATOM 22 CB GLN A 2 -9.270 7.360 2.255 1.00 0.00 C \nATOM 23 CG GLN A 2 -8.419 8.445 1.575 1.00 0.00 C \nATOM 24 CD GLN A 2 -8.134 9.580 2.563 1.00 0.00 C \nATOM 25 OE1 GLN A 2 -6.993 9.933 2.789 1.00 0.00 O \nATOM 26 NE2 GLN A 2 -9.131 10.170 3.164 1.00 0.00 N \nATOM 27 H GLN A 2 -11.278 6.556 3.474 1.00 0.00 H \nATOM 28 HA GLN A 2 -10.887 8.161 1.079 1.00 0.00 H \nATOM 29 HB2 GLN A 2 -9.448 7.628 3.287 1.00 0.00 H \nATOM 30 HB3 GLN A 2 -8.744 6.419 2.215 1.00 0.00 H \nATOM 31 HG2 GLN A 2 -7.484 8.012 1.249 1.00 0.00 H \nATOM 32 HG3 GLN A 2 -8.947 8.841 0.719 1.00 0.00 H \nATOM 33 HE21 GLN A 2 -10.051 9.886 2.982 1.00 0.00 H \nATOM 34 HE22 GLN A 2 -8.960 10.898 3.797 1.00 0.00 H \nATOM 35 N TYR A 3 -9.646 6.258 -0.481 1.00 0.00 N \nATOM 36 CA TYR A 3 -9.515 5.226 -1.559 1.00 0.00 C \nATOM 37 C TYR A 3 -9.305 3.829 -0.971 1.00 0.00 C \nATOM 38 O TYR A 3 -8.779 3.671 0.115 1.00 0.00 O \nATOM 39 CB TYR A 3 -8.310 5.647 -2.405 1.00 0.00 C \nATOM 40 CG TYR A 3 -7.109 5.903 -1.529 1.00 0.00 C \nATOM 41 CD1 TYR A 3 -6.471 4.847 -0.869 1.00 0.00 C \nATOM 42 CD2 TYR A 3 -6.643 7.209 -1.378 1.00 0.00 C \nATOM 43 CE1 TYR A 3 -5.364 5.106 -0.061 1.00 0.00 C \nATOM 44 CE2 TYR A 3 -5.538 7.467 -0.570 1.00 0.00 C \nATOM 45 CZ TYR A 3 -4.896 6.416 0.090 1.00 0.00 C \nATOM 46 OH TYR A 3 -3.803 6.671 0.889 1.00 0.00 O \nATOM 47 H TYR A 3 -9.063 7.046 -0.494 1.00 0.00 H \nATOM 48 HA TYR A 3 -10.391 5.232 -2.170 1.00 0.00 H \nATOM 49 HB2 TYR A 3 -8.077 4.866 -3.113 1.00 0.00 H \nATOM 50 HB3 TYR A 3 -8.557 6.552 -2.937 1.00 0.00 H \nATOM 51 HD1 TYR A 3 -6.829 3.833 -0.981 1.00 0.00 H \nATOM 52 HD2 TYR A 3 -7.137 8.022 -1.889 1.00 0.00 H \nATOM 53 HE1 TYR A 3 -4.873 4.297 0.447 1.00 0.00 H \nATOM 54 HE2 TYR A 3 -5.182 8.475 -0.454 1.00 0.00 H \nATOM 55 HH TYR A 3 -4.102 7.182 1.645 1.00 0.00 H \nATOM 56 N THR A 4 -9.719 2.816 -1.690 1.00 0.00 N \nATOM 57 CA THR A 4 -9.557 1.419 -1.196 1.00 0.00 C \nATOM 58 C THR A 4 -8.535 0.673 -2.059 1.00 0.00 C \nATOM 59 O THR A 4 -8.743 -0.461 -2.445 1.00 0.00 O \nATOM 60 CB THR A 4 -10.946 0.787 -1.336 1.00 0.00 C \nATOM 61 OG1 THR A 4 -11.486 1.125 -2.606 1.00 0.00 O \nATOM 62 CG2 THR A 4 -11.871 1.309 -0.232 1.00 0.00 C \nATOM 63 H THR A 4 -10.141 2.979 -2.559 1.00 0.00 H \nATOM 64 HA THR A 4 -9.253 1.416 -0.161 1.00 0.00 H \nATOM 65 HB THR A 4 -10.863 -0.285 -1.255 1.00 0.00 H \nATOM 66 HG1 THR A 4 -11.537 0.322 -3.130 1.00 0.00 H \nATOM 67 HG21 THR A 4 -11.313 1.936 0.448 1.00 0.00 H \nATOM 68 HG22 THR A 4 -12.289 0.474 0.311 1.00 0.00 H \nATOM 69 HG23 THR A 4 -12.671 1.884 -0.676 1.00 0.00 H \nATOM 70 N ALA A 5 -7.431 1.309 -2.365 1.00 0.00 N \nATOM 71 CA ALA A 5 -6.376 0.662 -3.210 1.00 0.00 C \nATOM 72 C ALA A 5 -6.018 -0.724 -2.683 1.00 0.00 C \nATOM 73 O ALA A 5 -5.727 -0.879 -1.525 1.00 0.00 O \nATOM 74 CB ALA A 5 -5.150 1.563 -3.080 1.00 0.00 C \nATOM 75 H ALA A 5 -7.297 2.222 -2.044 1.00 0.00 H \nATOM 76 HA ALA A 5 -6.690 0.615 -4.236 1.00 0.00 H \nATOM 77 HB1 ALA A 5 -5.459 2.557 -2.801 1.00 0.00 H \nATOM 78 HB2 ALA A 5 -4.628 1.595 -4.025 1.00 0.00 H \nATOM 79 HB3 ALA A 5 -4.489 1.159 -2.316 1.00 0.00 H \nATOM 80 N LYS A 6 -6.002 -1.715 -3.525 1.00 0.00 N \nATOM 81 CA LYS A 6 -5.621 -3.082 -3.057 1.00 0.00 C \nATOM 82 C LYS A 6 -4.348 -3.527 -3.770 1.00 0.00 C \nATOM 83 O LYS A 6 -4.149 -3.252 -4.939 1.00 0.00 O \nATOM 84 CB LYS A 6 -6.793 -4.004 -3.395 1.00 0.00 C \nATOM 85 CG LYS A 6 -7.202 -3.833 -4.861 1.00 0.00 C \nATOM 86 CD LYS A 6 -7.597 -5.193 -5.460 1.00 0.00 C \nATOM 87 CE LYS A 6 -9.078 -5.181 -5.849 1.00 0.00 C \nATOM 88 NZ LYS A 6 -9.092 -4.834 -7.297 1.00 0.00 N \nATOM 89 H LYS A 6 -6.215 -1.559 -4.467 1.00 0.00 H \nATOM 90 HA LYS A 6 -5.460 -3.073 -1.990 1.00 0.00 H \nATOM 91 HB2 LYS A 6 -6.496 -5.028 -3.222 1.00 0.00 H \nATOM 92 HB3 LYS A 6 -7.631 -3.760 -2.759 1.00 0.00 H \nATOM 93 HG2 LYS A 6 -8.038 -3.152 -4.916 1.00 0.00 H \nATOM 94 HG3 LYS A 6 -6.371 -3.426 -5.416 1.00 0.00 H \nATOM 95 HD2 LYS A 6 -6.997 -5.383 -6.339 1.00 0.00 H \nATOM 96 HD3 LYS A 6 -7.425 -5.975 -4.734 1.00 0.00 H \nATOM 97 HE2 LYS A 6 -9.516 -6.157 -5.690 1.00 0.00 H \nATOM 98 HE3 LYS A 6 -9.611 -4.431 -5.284 1.00 0.00 H \nATOM 99 HZ1 LYS A 6 -10.075 -4.762 -7.627 1.00 0.00 H \nATOM 100 HZ2 LYS A 6 -8.597 -5.574 -7.835 1.00 0.00 H \nATOM 101 HZ3 LYS A 6 -8.613 -3.922 -7.440 1.00 0.00 H \nATOM 102 N TYR A 7 -3.479 -4.197 -3.064 1.00 0.00 N \nATOM 103 CA TYR A 7 -2.198 -4.652 -3.679 1.00 0.00 C \nATOM 104 C TYR A 7 -2.015 -6.158 -3.475 1.00 0.00 C \nATOM 105 O TYR A 7 -2.084 -6.931 -4.412 1.00 0.00 O \nATOM 106 CB TYR A 7 -1.117 -3.858 -2.948 1.00 0.00 C \nATOM 107 CG TYR A 7 -1.118 -2.455 -3.461 1.00 0.00 C \nATOM 108 CD1 TYR A 7 -2.152 -1.589 -3.106 1.00 0.00 C \nATOM 109 CD2 TYR A 7 -0.084 -2.022 -4.286 1.00 0.00 C \nATOM 110 CE1 TYR A 7 -2.155 -0.278 -3.578 1.00 0.00 C \nATOM 111 CE2 TYR A 7 -0.076 -0.713 -4.762 1.00 0.00 C \nATOM 112 CZ TYR A 7 -1.113 0.167 -4.409 1.00 0.00 C \nATOM 113 OH TYR A 7 -1.107 1.464 -4.879 1.00 0.00 O \nATOM 114 H TYR A 7 -3.666 -4.392 -2.123 1.00 0.00 H \nATOM 115 HA TYR A 7 -2.179 -4.405 -4.728 1.00 0.00 H \nATOM 116 HB2 TYR A 7 -1.318 -3.841 -1.890 1.00 0.00 H \nATOM 117 HB3 TYR A 7 -0.154 -4.299 -3.125 1.00 0.00 H \nATOM 118 HD1 TYR A 7 -2.949 -1.935 -2.468 1.00 0.00 H \nATOM 119 HD2 TYR A 7 0.709 -2.702 -4.555 1.00 0.00 H \nATOM 120 HE1 TYR A 7 -2.953 0.398 -3.287 1.00 0.00 H \nATOM 121 HE2 TYR A 7 0.731 -0.383 -5.395 1.00 0.00 H \nATOM 122 HH TYR A 7 -2.007 1.693 -5.125 1.00 0.00 H \nATOM 123 N LYS A 8 -1.794 -6.578 -2.256 1.00 0.00 N \nATOM 124 CA LYS A 8 -1.617 -8.034 -1.979 1.00 0.00 C \nATOM 125 C LYS A 8 -2.715 -8.510 -1.023 1.00 0.00 C \nATOM 126 O LYS A 8 -2.443 -8.993 0.061 1.00 0.00 O \nATOM 127 CB LYS A 8 -0.240 -8.151 -1.323 1.00 0.00 C \nATOM 128 CG LYS A 8 0.808 -8.472 -2.391 1.00 0.00 C \nATOM 129 CD LYS A 8 0.652 -9.925 -2.842 1.00 0.00 C \nATOM 130 CE LYS A 8 1.289 -10.854 -1.806 1.00 0.00 C \nATOM 131 NZ LYS A 8 0.671 -12.187 -2.052 1.00 0.00 N \nATOM 132 H LYS A 8 -1.751 -5.933 -1.520 1.00 0.00 H \nATOM 133 HA LYS A 8 -1.642 -8.600 -2.896 1.00 0.00 H \nATOM 134 HB2 LYS A 8 0.010 -7.217 -0.841 1.00 0.00 H \nATOM 135 HB3 LYS A 8 -0.256 -8.943 -0.590 1.00 0.00 H \nATOM 136 HG2 LYS A 8 0.671 -7.814 -3.238 1.00 0.00 H \nATOM 137 HG3 LYS A 8 1.796 -8.328 -1.980 1.00 0.00 H \nATOM 138 HD2 LYS A 8 -0.398 -10.160 -2.942 1.00 0.00 H \nATOM 139 HD3 LYS A 8 1.144 -10.062 -3.794 1.00 0.00 H \nATOM 140 HE2 LYS A 8 2.359 -10.900 -1.951 1.00 0.00 H \nATOM 141 HE3 LYS A 8 1.057 -10.518 -0.807 1.00 0.00 H \nATOM 142 HZ1 LYS A 8 -0.361 -12.117 -1.946 1.00 0.00 H \nATOM 143 HZ2 LYS A 8 1.045 -12.874 -1.365 1.00 0.00 H \nATOM 144 HZ3 LYS A 8 0.898 -12.502 -3.016 1.00 0.00 H \nATOM 145 N GLY A 9 -3.954 -8.363 -1.417 1.00 0.00 N \nATOM 146 CA GLY A 9 -5.080 -8.789 -0.537 1.00 0.00 C \nATOM 147 C GLY A 9 -5.175 -7.832 0.651 1.00 0.00 C \nATOM 148 O GLY A 9 -5.590 -8.208 1.731 1.00 0.00 O \nATOM 149 H GLY A 9 -4.142 -7.963 -2.292 1.00 0.00 H \nATOM 150 HA2 GLY A 9 -6.004 -8.766 -1.099 1.00 0.00 H \nATOM 151 HA3 GLY A 9 -4.899 -9.790 -0.177 1.00 0.00 H \nATOM 152 N ARG A 10 -4.785 -6.595 0.458 1.00 0.00 N \nATOM 153 CA ARG A 10 -4.842 -5.602 1.571 1.00 0.00 C \nATOM 154 C ARG A 10 -5.215 -4.223 1.028 1.00 0.00 C \nATOM 155 O ARG A 10 -4.428 -3.589 0.348 1.00 0.00 O \nATOM 156 CB ARG A 10 -3.424 -5.563 2.144 1.00 0.00 C \nATOM 157 CG ARG A 10 -3.028 -6.951 2.654 1.00 0.00 C \nATOM 158 CD ARG A 10 -1.692 -6.857 3.394 1.00 0.00 C \nATOM 159 NE ARG A 10 -1.225 -8.264 3.530 1.00 0.00 N \nATOM 160 CZ ARG A 10 -0.891 -8.731 4.702 1.00 0.00 C \nATOM 161 NH1 ARG A 10 0.174 -8.280 5.307 1.00 0.00 N \nATOM 162 NH2 ARG A 10 -1.622 -9.653 5.268 1.00 0.00 N \nATOM 163 H ARG A 10 -4.452 -6.321 -0.420 1.00 0.00 H \nATOM 164 HA ARG A 10 -5.541 -5.915 2.329 1.00 0.00 H \nATOM 165 HB2 ARG A 10 -2.733 -5.250 1.371 1.00 0.00 H \nATOM 166 HB3 ARG A 10 -3.387 -4.859 2.962 1.00 0.00 H \nATOM 167 HG2 ARG A 10 -3.790 -7.318 3.326 1.00 0.00 H \nATOM 168 HG3 ARG A 10 -2.928 -7.627 1.819 1.00 0.00 H \nATOM 169 HD2 ARG A 10 -0.984 -6.279 2.816 1.00 0.00 H \nATOM 170 HD3 ARG A 10 -1.831 -6.418 4.369 1.00 0.00 H \nATOM 171 HE ARG A 10 -1.167 -8.838 2.739 1.00 0.00 H \nATOM 172 HH11 ARG A 10 0.735 -7.576 4.873 1.00 0.00 H \nATOM 173 HH12 ARG A 10 0.428 -8.640 6.205 1.00 0.00 H \nATOM 174 HH21 ARG A 10 -2.437 -10.000 4.804 1.00 0.00 H \nATOM 175 HH22 ARG A 10 -1.366 -10.012 6.166 1.00 0.00 H \nATOM 176 N THR A 11 -6.396 -3.744 1.330 1.00 0.00 N \nATOM 177 CA THR A 11 -6.793 -2.394 0.833 1.00 0.00 C \nATOM 178 C THR A 11 -5.968 -1.326 1.561 1.00 0.00 C \nATOM 179 O THR A 11 -5.609 -1.495 2.712 1.00 0.00 O \nATOM 180 CB THR A 11 -8.276 -2.240 1.172 1.00 0.00 C \nATOM 181 OG1 THR A 11 -9.006 -3.320 0.606 1.00 0.00 O \nATOM 182 CG2 THR A 11 -8.791 -0.914 0.601 1.00 0.00 C \nATOM 183 H THR A 11 -7.011 -4.266 1.887 1.00 0.00 H \nATOM 184 HA THR A 11 -6.651 -2.336 -0.237 1.00 0.00 H \nATOM 185 HB THR A 11 -8.404 -2.240 2.244 1.00 0.00 H \nATOM 186 HG1 THR A 11 -9.706 -3.559 1.217 1.00 0.00 H \nATOM 187 HG21 THR A 11 -9.487 -0.470 1.297 1.00 0.00 H \nATOM 188 HG22 THR A 11 -9.287 -1.097 -0.339 1.00 0.00 H \nATOM 189 HG23 THR A 11 -7.959 -0.238 0.443 1.00 0.00 H \nATOM 190 N PHE A 12 -5.665 -0.234 0.908 1.00 0.00 N \nATOM 191 CA PHE A 12 -4.864 0.839 1.567 1.00 0.00 C \nATOM 192 C PHE A 12 -5.720 2.089 1.765 1.00 0.00 C \nATOM 193 O PHE A 12 -6.276 2.630 0.829 1.00 0.00 O \nATOM 194 CB PHE A 12 -3.688 1.110 0.620 1.00 0.00 C \nATOM 195 CG PHE A 12 -2.703 -0.015 0.763 1.00 0.00 C \nATOM 196 CD1 PHE A 12 -2.930 -1.198 0.069 1.00 0.00 C \nATOM 197 CD2 PHE A 12 -1.572 0.121 1.579 1.00 0.00 C \nATOM 198 CE1 PHE A 12 -2.031 -2.263 0.188 1.00 0.00 C \nATOM 199 CE2 PHE A 12 -0.666 -0.942 1.697 1.00 0.00 C \nATOM 200 CZ PHE A 12 -0.899 -2.136 1.003 1.00 0.00 C \nATOM 201 H PHE A 12 -5.965 -0.120 -0.018 1.00 0.00 H \nATOM 202 HA PHE A 12 -4.490 0.490 2.517 1.00 0.00 H \nATOM 203 HB2 PHE A 12 -4.035 1.151 -0.411 1.00 0.00 H \nATOM 204 HB3 PHE A 12 -3.214 2.042 0.885 1.00 0.00 H \nATOM 205 HD1 PHE A 12 -3.801 -1.281 -0.568 1.00 0.00 H \nATOM 206 HD2 PHE A 12 -1.401 1.040 2.120 1.00 0.00 H \nATOM 207 HE1 PHE A 12 -2.217 -3.186 -0.337 1.00 0.00 H \nATOM 208 HE2 PHE A 12 0.215 -0.840 2.313 1.00 0.00 H \nATOM 209 HZ PHE A 12 -0.203 -2.957 1.091 1.00 0.00 H \nATOM 210 N ARG A 13 -5.832 2.540 2.986 1.00 0.00 N \nATOM 211 CA ARG A 13 -6.647 3.755 3.277 1.00 0.00 C \nATOM 212 C ARG A 13 -5.750 4.832 3.885 1.00 0.00 C \nATOM 213 O ARG A 13 -6.164 5.593 4.739 1.00 0.00 O \nATOM 214 CB ARG A 13 -7.697 3.294 4.289 1.00 0.00 C \nATOM 215 CG ARG A 13 -9.002 4.058 4.057 1.00 0.00 C \nATOM 216 CD ARG A 13 -9.889 3.271 3.090 1.00 0.00 C \nATOM 217 NE ARG A 13 -10.468 2.171 3.910 1.00 0.00 N \nATOM 218 CZ ARG A 13 -11.553 2.376 4.605 1.00 0.00 C \nATOM 219 NH1 ARG A 13 -12.723 2.235 4.045 1.00 0.00 N \nATOM 220 NH2 ARG A 13 -11.468 2.722 5.862 1.00 0.00 N \nATOM 221 H ARG A 13 -5.375 2.077 3.719 1.00 0.00 H \nATOM 222 HA ARG A 13 -7.125 4.116 2.381 1.00 0.00 H \nATOM 223 HB2 ARG A 13 -7.873 2.234 4.168 1.00 0.00 H \nATOM 224 HB3 ARG A 13 -7.342 3.489 5.290 1.00 0.00 H \nATOM 225 HG2 ARG A 13 -9.516 4.185 4.999 1.00 0.00 H \nATOM 226 HG3 ARG A 13 -8.781 5.026 3.634 1.00 0.00 H \nATOM 227 HD2 ARG A 13 -10.674 3.906 2.701 1.00 0.00 H \nATOM 228 HD3 ARG A 13 -9.299 2.862 2.285 1.00 0.00 H \nATOM 229 HE ARG A 13 -10.033 1.293 3.929 1.00 0.00 H \nATOM 230 HH11 ARG A 13 -12.788 1.970 3.083 1.00 0.00 H \nATOM 231 HH12 ARG A 13 -13.555 2.393 4.577 1.00 0.00 H \nATOM 232 HH21 ARG A 13 -10.571 2.829 6.290 1.00 0.00 H \nATOM 233 HH22 ARG A 13 -12.299 2.879 6.394 1.00 0.00 H \nATOM 234 N ASN A 14 -4.519 4.889 3.449 1.00 0.00 N \nATOM 235 CA ASN A 14 -3.569 5.905 3.992 1.00 0.00 C \nATOM 236 C ASN A 14 -2.334 5.998 3.094 1.00 0.00 C \nATOM 237 O ASN A 14 -1.704 5.003 2.785 1.00 0.00 O \nATOM 238 CB ASN A 14 -3.188 5.383 5.378 1.00 0.00 C \nATOM 239 CG ASN A 14 -3.025 6.559 6.341 1.00 0.00 C \nATOM 240 OD1 ASN A 14 -3.961 6.946 7.011 1.00 0.00 O \nATOM 241 ND2 ASN A 14 -1.866 7.149 6.438 1.00 0.00 N \nATOM 242 H ASN A 14 -4.217 4.258 2.760 1.00 0.00 H \nATOM 243 HA ASN A 14 -4.050 6.866 4.078 1.00 0.00 H \nATOM 244 HB2 ASN A 14 -3.964 4.724 5.741 1.00 0.00 H \nATOM 245 HB3 ASN A 14 -2.256 4.841 5.315 1.00 0.00 H \nATOM 246 HD21 ASN A 14 -1.111 6.836 5.898 1.00 0.00 H \nATOM 247 HD22 ASN A 14 -1.750 7.903 7.052 1.00 0.00 H \nATOM 248 N GLU A 15 -1.991 7.185 2.667 1.00 0.00 N \nATOM 249 CA GLU A 15 -0.800 7.354 1.778 1.00 0.00 C \nATOM 250 C GLU A 15 0.469 6.873 2.490 1.00 0.00 C \nATOM 251 O GLU A 15 1.371 6.340 1.872 1.00 0.00 O \nATOM 252 CB GLU A 15 -0.721 8.855 1.489 1.00 0.00 C \nATOM 253 CG GLU A 15 -0.269 9.077 0.045 1.00 0.00 C \nATOM 254 CD GLU A 15 0.523 10.382 -0.048 1.00 0.00 C \nATOM 255 OE1 GLU A 15 1.362 10.603 0.809 1.00 0.00 O \nATOM 256 OE2 GLU A 15 0.279 11.135 -0.975 1.00 0.00 O \nATOM 257 H GLU A 15 -2.522 7.968 2.928 1.00 0.00 H \nATOM 258 HA GLU A 15 -0.941 6.811 0.857 1.00 0.00 H \nATOM 259 HB2 GLU A 15 -1.697 9.300 1.634 1.00 0.00 H \nATOM 260 HB3 GLU A 15 -0.012 9.315 2.162 1.00 0.00 H \nATOM 261 HG2 GLU A 15 0.354 8.251 -0.267 1.00 0.00 H \nATOM 262 HG3 GLU A 15 -1.135 9.137 -0.598 1.00 0.00 H \nATOM 263 N LYS A 16 0.545 7.060 3.784 1.00 0.00 N \nATOM 264 CA LYS A 16 1.753 6.619 4.545 1.00 0.00 C \nATOM 265 C LYS A 16 1.980 5.119 4.366 1.00 0.00 C \nATOM 266 O LYS A 16 3.094 4.656 4.212 1.00 0.00 O \nATOM 267 CB LYS A 16 1.451 6.940 6.009 1.00 0.00 C \nATOM 268 CG LYS A 16 2.737 6.830 6.832 1.00 0.00 C \nATOM 269 CD LYS A 16 2.398 6.393 8.259 1.00 0.00 C \nATOM 270 CE LYS A 16 3.484 5.447 8.776 1.00 0.00 C \nATOM 271 NZ LYS A 16 2.754 4.433 9.586 1.00 0.00 N \nATOM 272 H LYS A 16 -0.192 7.495 4.255 1.00 0.00 H \nATOM 273 HA LYS A 16 2.607 7.164 4.220 1.00 0.00 H \nATOM 274 HB2 LYS A 16 1.058 7.943 6.085 1.00 0.00 H \nATOM 275 HB3 LYS A 16 0.724 6.237 6.387 1.00 0.00 H \nATOM 276 HG2 LYS A 16 3.394 6.102 6.376 1.00 0.00 H \nATOM 277 HG3 LYS A 16 3.229 7.791 6.858 1.00 0.00 H \nATOM 278 HD2 LYS A 16 2.344 7.263 8.898 1.00 0.00 H \nATOM 279 HD3 LYS A 16 1.446 5.882 8.265 1.00 0.00 H \nATOM 280 HE2 LYS A 16 3.994 4.974 7.948 1.00 0.00 H \nATOM 281 HE3 LYS A 16 4.185 5.981 9.398 1.00 0.00 H \nATOM 282 HZ1 LYS A 16 3.417 3.699 9.906 1.00 0.00 H \nATOM 283 HZ2 LYS A 16 2.008 3.998 9.005 1.00 0.00 H \nATOM 284 HZ3 LYS A 16 2.325 4.892 10.414 1.00 0.00 H \nATOM 285 N GLU A 17 0.920 4.370 4.386 1.00 0.00 N \nATOM 286 CA GLU A 17 1.030 2.887 4.218 1.00 0.00 C \nATOM 287 C GLU A 17 1.435 2.550 2.782 1.00 0.00 C \nATOM 288 O GLU A 17 2.432 1.897 2.540 1.00 0.00 O \nATOM 289 CB GLU A 17 -0.372 2.347 4.510 1.00 0.00 C \nATOM 290 CG GLU A 17 -0.665 2.465 6.005 1.00 0.00 C \nATOM 291 CD GLU A 17 0.266 1.532 6.784 1.00 0.00 C \nATOM 292 OE1 GLU A 17 0.483 0.423 6.324 1.00 0.00 O \nATOM 293 OE2 GLU A 17 0.746 1.944 7.827 1.00 0.00 O \nATOM 294 H GLU A 17 0.047 4.789 4.511 1.00 0.00 H \nATOM 295 HA GLU A 17 1.739 2.476 4.919 1.00 0.00 H \nATOM 296 HB2 GLU A 17 -1.100 2.920 3.954 1.00 0.00 H \nATOM 297 HB3 GLU A 17 -0.428 1.311 4.214 1.00 0.00 H \nATOM 298 HG2 GLU A 17 -0.504 3.485 6.323 1.00 0.00 H \nATOM 299 HG3 GLU A 17 -1.690 2.185 6.192 1.00 0.00 H \nATOM 300 N LEU A 18 0.656 2.993 1.830 1.00 0.00 N \nATOM 301 CA LEU A 18 0.963 2.712 0.391 1.00 0.00 C \nATOM 302 C LEU A 18 2.375 3.187 0.038 1.00 0.00 C \nATOM 303 O LEU A 18 3.118 2.493 -0.632 1.00 0.00 O \nATOM 304 CB LEU A 18 -0.099 3.496 -0.395 1.00 0.00 C \nATOM 305 CG LEU A 18 -0.416 2.797 -1.723 1.00 0.00 C \nATOM 306 CD1 LEU A 18 -0.950 1.376 -1.466 1.00 0.00 C \nATOM 307 CD2 LEU A 18 -1.476 3.609 -2.475 1.00 0.00 C \nATOM 308 H LEU A 18 -0.142 3.511 2.064 1.00 0.00 H \nATOM 309 HA LEU A 18 0.872 1.659 0.193 1.00 0.00 H \nATOM 310 HB2 LEU A 18 -1.000 3.565 0.196 1.00 0.00 H \nATOM 311 HB3 LEU A 18 0.271 4.491 -0.597 1.00 0.00 H \nATOM 312 HG LEU A 18 0.478 2.747 -2.318 1.00 0.00 H \nATOM 313 HD11 LEU A 18 -0.814 1.123 -0.438 1.00 0.00 H \nATOM 314 HD12 LEU A 18 -0.413 0.662 -2.077 1.00 0.00 H \nATOM 315 HD13 LEU A 18 -2.001 1.331 -1.706 1.00 0.00 H \nATOM 316 HD21 LEU A 18 -2.346 3.735 -1.847 1.00 0.00 H \nATOM 317 HD22 LEU A 18 -1.756 3.087 -3.378 1.00 0.00 H \nATOM 318 HD23 LEU A 18 -1.073 4.578 -2.729 1.00 0.00 H \nATOM 319 N ARG A 19 2.756 4.351 0.493 1.00 0.00 N \nATOM 320 CA ARG A 19 4.129 4.857 0.190 1.00 0.00 C \nATOM 321 C ARG A 19 5.183 3.920 0.792 1.00 0.00 C \nATOM 322 O ARG A 19 6.327 3.919 0.379 1.00 0.00 O \nATOM 323 CB ARG A 19 4.206 6.239 0.841 1.00 0.00 C \nATOM 324 CG ARG A 19 3.398 7.243 0.014 1.00 0.00 C \nATOM 325 CD ARG A 19 4.332 7.990 -0.940 1.00 0.00 C \nATOM 326 NE ARG A 19 3.492 8.312 -2.126 1.00 0.00 N \nATOM 327 CZ ARG A 19 3.911 8.009 -3.324 1.00 0.00 C \nATOM 328 NH1 ARG A 19 4.462 6.846 -3.549 1.00 0.00 N \nATOM 329 NH2 ARG A 19 3.781 8.868 -4.297 1.00 0.00 N \nATOM 330 H ARG A 19 2.142 4.886 1.039 1.00 0.00 H \nATOM 331 HA ARG A 19 4.270 4.944 -0.874 1.00 0.00 H \nATOM 332 HB2 ARG A 19 3.803 6.187 1.842 1.00 0.00 H \nATOM 333 HB3 ARG A 19 5.236 6.557 0.883 1.00 0.00 H \nATOM 334 HG2 ARG A 19 2.644 6.717 -0.555 1.00 0.00 H \nATOM 335 HG3 ARG A 19 2.920 7.951 0.675 1.00 0.00 H \nATOM 336 HD2 ARG A 19 4.693 8.898 -0.477 1.00 0.00 H \nATOM 337 HD3 ARG A 19 5.156 7.360 -1.231 1.00 0.00 H \nATOM 338 HE ARG A 19 2.627 8.753 -2.007 1.00 0.00 H \nATOM 339 HH11 ARG A 19 4.562 6.188 -2.802 1.00 0.00 H \nATOM 340 HH12 ARG A 19 4.782 6.614 -4.467 1.00 0.00 H \nATOM 341 HH21 ARG A 19 3.361 9.759 -4.125 1.00 0.00 H \nATOM 342 HH22 ARG A 19 4.100 8.636 -5.216 1.00 0.00 H \nATOM 343 N ASP A 20 4.807 3.123 1.766 1.00 0.00 N \nATOM 344 CA ASP A 20 5.785 2.188 2.395 1.00 0.00 C \nATOM 345 C ASP A 20 5.599 0.769 1.846 1.00 0.00 C \nATOM 346 O ASP A 20 6.535 -0.006 1.791 1.00 0.00 O \nATOM 347 CB ASP A 20 5.466 2.230 3.890 1.00 0.00 C \nATOM 348 CG ASP A 20 6.623 1.613 4.678 1.00 0.00 C \nATOM 349 OD1 ASP A 20 7.761 1.903 4.347 1.00 0.00 O \nATOM 350 OD2 ASP A 20 6.351 0.862 5.600 1.00 0.00 O \nATOM 351 H ASP A 20 3.882 3.142 2.086 1.00 0.00 H \nATOM 352 HA ASP A 20 6.794 2.527 2.225 1.00 0.00 H \nATOM 353 HB2 ASP A 20 5.325 3.256 4.199 1.00 0.00 H \nATOM 354 HB3 ASP A 20 4.563 1.669 4.082 1.00 0.00 H \nATOM 355 N PHE A 21 4.399 0.422 1.443 1.00 0.00 N \nATOM 356 CA PHE A 21 4.159 -0.951 0.898 1.00 0.00 C \nATOM 357 C PHE A 21 4.866 -1.132 -0.452 1.00 0.00 C \nATOM 358 O PHE A 21 5.866 -1.815 -0.552 1.00 0.00 O \nATOM 359 CB PHE A 21 2.644 -1.080 0.725 1.00 0.00 C \nATOM 360 CG PHE A 21 2.364 -2.455 0.180 1.00 0.00 C \nATOM 361 CD1 PHE A 21 2.350 -3.536 1.053 1.00 0.00 C \nATOM 362 CD2 PHE A 21 2.149 -2.647 -1.189 1.00 0.00 C \nATOM 363 CE1 PHE A 21 2.110 -4.827 0.567 1.00 0.00 C \nATOM 364 CE2 PHE A 21 1.910 -3.935 -1.681 1.00 0.00 C \nATOM 365 CZ PHE A 21 1.887 -5.027 -0.801 1.00 0.00 C \nATOM 366 H PHE A 21 3.659 1.063 1.498 1.00 0.00 H \nATOM 367 HA PHE A 21 4.496 -1.702 1.602 1.00 0.00 H \nATOM 368 HB2 PHE A 21 2.165 -0.961 1.678 1.00 0.00 H \nATOM 369 HB3 PHE A 21 2.271 -0.334 0.047 1.00 0.00 H \nATOM 370 HD1 PHE A 21 2.538 -3.370 2.103 1.00 0.00 H \nATOM 371 HD2 PHE A 21 2.171 -1.802 -1.867 1.00 0.00 H \nATOM 372 HE1 PHE A 21 2.093 -5.666 1.247 1.00 0.00 H \nATOM 373 HE2 PHE A 21 1.748 -4.089 -2.736 1.00 0.00 H \nATOM 374 HZ PHE A 21 1.702 -6.021 -1.179 1.00 0.00 H \nATOM 375 N ILE A 22 4.332 -0.536 -1.495 1.00 0.00 N \nATOM 376 CA ILE A 22 4.937 -0.668 -2.861 1.00 0.00 C \nATOM 377 C ILE A 22 6.448 -0.403 -2.800 1.00 0.00 C \nATOM 378 O ILE A 22 7.225 -0.980 -3.537 1.00 0.00 O \nATOM 379 CB ILE A 22 4.230 0.391 -3.714 1.00 0.00 C \nATOM 380 CG1 ILE A 22 2.755 0.011 -3.863 1.00 0.00 C \nATOM 381 CG2 ILE A 22 4.863 0.434 -5.107 1.00 0.00 C \nATOM 382 CD1 ILE A 22 1.887 0.886 -2.968 1.00 0.00 C \nATOM 383 H ILE A 22 3.521 -0.008 -1.377 1.00 0.00 H \nATOM 384 HA ILE A 22 4.730 -1.648 -3.270 1.00 0.00 H \nATOM 385 HB ILE A 22 4.317 1.359 -3.242 1.00 0.00 H \nATOM 386 HG12 ILE A 22 2.455 0.145 -4.888 1.00 0.00 H \nATOM 387 HG13 ILE A 22 2.622 -1.020 -3.582 1.00 0.00 H \nATOM 388 HG21 ILE A 22 4.292 1.096 -5.739 1.00 0.00 H \nATOM 389 HG22 ILE A 22 4.857 -0.560 -5.525 1.00 0.00 H \nATOM 390 HG23 ILE A 22 5.879 0.790 -5.030 1.00 0.00 H \nATOM 391 HD11 ILE A 22 1.555 0.309 -2.116 1.00 0.00 H \nATOM 392 HD12 ILE A 22 1.027 1.226 -3.526 1.00 0.00 H \nATOM 393 HD13 ILE A 22 2.456 1.737 -2.628 1.00 0.00 H \nATOM 394 N GLU A 23 6.851 0.463 -1.911 1.00 0.00 N \nATOM 395 CA GLU A 23 8.304 0.776 -1.772 1.00 0.00 C \nATOM 396 C GLU A 23 9.036 -0.436 -1.197 1.00 0.00 C \nATOM 397 O GLU A 23 10.184 -0.688 -1.516 1.00 0.00 O \nATOM 398 CB GLU A 23 8.372 1.958 -0.802 1.00 0.00 C \nATOM 399 CG GLU A 23 9.792 2.527 -0.785 1.00 0.00 C \nATOM 400 CD GLU A 23 9.846 3.737 0.148 1.00 0.00 C \nATOM 401 OE1 GLU A 23 9.504 3.580 1.309 1.00 0.00 O \nATOM 402 OE2 GLU A 23 10.227 4.800 -0.313 1.00 0.00 O \nATOM 403 H GLU A 23 6.190 0.901 -1.329 1.00 0.00 H \nATOM 404 HA GLU A 23 8.724 1.053 -2.725 1.00 0.00 H \nATOM 405 HB2 GLU A 23 7.679 2.724 -1.120 1.00 0.00 H \nATOM 406 HB3 GLU A 23 8.108 1.624 0.191 1.00 0.00 H \nATOM 407 HG2 GLU A 23 10.478 1.770 -0.435 1.00 0.00 H \nATOM 408 HG3 GLU A 23 10.070 2.832 -1.783 1.00 0.00 H \nATOM 409 N LYS A 24 8.373 -1.193 -0.361 1.00 0.00 N \nATOM 410 CA LYS A 24 9.014 -2.402 0.232 1.00 0.00 C \nATOM 411 C LYS A 24 8.798 -3.604 -0.687 1.00 0.00 C \nATOM 412 O LYS A 24 9.740 -4.241 -1.117 1.00 0.00 O \nATOM 413 CB LYS A 24 8.309 -2.613 1.573 1.00 0.00 C \nATOM 414 CG LYS A 24 9.198 -2.096 2.708 1.00 0.00 C \nATOM 415 CD LYS A 24 10.424 -3.005 2.855 1.00 0.00 C \nATOM 416 CE LYS A 24 11.664 -2.155 3.138 1.00 0.00 C \nATOM 417 NZ LYS A 24 11.492 -1.677 4.538 1.00 0.00 N \nATOM 418 H LYS A 24 7.447 -0.970 -0.130 1.00 0.00 H \nATOM 419 HA LYS A 24 10.068 -2.232 0.388 1.00 0.00 H \nATOM 420 HB2 LYS A 24 7.371 -2.075 1.576 1.00 0.00 H \nATOM 421 HB3 LYS A 24 8.119 -3.666 1.719 1.00 0.00 H \nATOM 422 HG2 LYS A 24 9.517 -1.089 2.483 1.00 0.00 H \nATOM 423 HG3 LYS A 24 8.638 -2.099 3.631 1.00 0.00 H \nATOM 424 HD2 LYS A 24 10.264 -3.692 3.674 1.00 0.00 H \nATOM 425 HD3 LYS A 24 10.573 -3.563 1.943 1.00 0.00 H \nATOM 426 HE2 LYS A 24 12.559 -2.756 3.050 1.00 0.00 H \nATOM 427 HE3 LYS A 24 11.707 -1.314 2.463 1.00 0.00 H \nATOM 428 HZ1 LYS A 24 10.620 -1.117 4.609 1.00 0.00 H \nATOM 429 HZ2 LYS A 24 12.307 -1.087 4.804 1.00 0.00 H \nATOM 430 HZ3 LYS A 24 11.431 -2.494 5.178 1.00 0.00 H \nATOM 431 N PHE A 25 7.561 -3.914 -0.997 1.00 0.00 N \nATOM 432 CA PHE A 25 7.279 -5.066 -1.891 1.00 0.00 C \nATOM 433 C PHE A 25 7.367 -4.635 -3.359 1.00 0.00 C \nATOM 434 O PHE A 25 6.438 -4.814 -4.124 1.00 0.00 O \nATOM 435 CB PHE A 25 5.858 -5.511 -1.541 1.00 0.00 C \nATOM 436 CG PHE A 25 5.548 -6.806 -2.249 1.00 0.00 C \nATOM 437 CD1 PHE A 25 6.396 -7.911 -2.098 1.00 0.00 C \nATOM 438 CD2 PHE A 25 4.411 -6.902 -3.059 1.00 0.00 C \nATOM 439 CE1 PHE A 25 6.105 -9.111 -2.758 1.00 0.00 C \nATOM 440 CE2 PHE A 25 4.121 -8.102 -3.719 1.00 0.00 C \nATOM 441 CZ PHE A 25 4.967 -9.207 -3.568 1.00 0.00 C \nATOM 442 H PHE A 25 6.822 -3.389 -0.641 1.00 0.00 H \nATOM 443 HA PHE A 25 7.969 -5.858 -1.688 1.00 0.00 H \nATOM 444 HB2 PHE A 25 5.777 -5.654 -0.473 1.00 0.00 H \nATOM 445 HB3 PHE A 25 5.155 -4.753 -1.857 1.00 0.00 H \nATOM 446 HD1 PHE A 25 7.274 -7.835 -1.475 1.00 0.00 H \nATOM 447 HD2 PHE A 25 3.760 -6.048 -3.173 1.00 0.00 H \nATOM 448 HE1 PHE A 25 6.758 -9.964 -2.642 1.00 0.00 H \nATOM 449 HE2 PHE A 25 3.243 -8.175 -4.343 1.00 0.00 H \nATOM 450 HZ PHE A 25 4.743 -10.132 -4.077 1.00 0.00 H \nATOM 451 N LYS A 26 8.480 -4.069 -3.753 1.00 0.00 N \nATOM 452 CA LYS A 26 8.638 -3.621 -5.173 1.00 0.00 C \nATOM 453 C LYS A 26 8.589 -4.815 -6.130 1.00 0.00 C \nATOM 454 O LYS A 26 8.310 -4.667 -7.304 1.00 0.00 O \nATOM 455 CB LYS A 26 10.009 -2.945 -5.233 1.00 0.00 C \nATOM 456 CG LYS A 26 9.861 -1.461 -4.890 1.00 0.00 C \nATOM 457 CD LYS A 26 11.141 -0.719 -5.279 1.00 0.00 C \nATOM 458 CE LYS A 26 11.003 -0.172 -6.701 1.00 0.00 C \nATOM 459 NZ LYS A 26 12.333 0.423 -7.013 1.00 0.00 N \nATOM 460 H LYS A 26 9.210 -3.937 -3.115 1.00 0.00 H \nATOM 461 HA LYS A 26 7.871 -2.917 -5.424 1.00 0.00 H \nATOM 462 HB2 LYS A 26 10.672 -3.415 -4.522 1.00 0.00 H \nATOM 463 HB3 LYS A 26 10.417 -3.045 -6.227 1.00 0.00 H \nATOM 464 HG2 LYS A 26 9.024 -1.048 -5.435 1.00 0.00 H \nATOM 465 HG3 LYS A 26 9.691 -1.351 -3.830 1.00 0.00 H \nATOM 466 HD2 LYS A 26 11.305 0.099 -4.592 1.00 0.00 H \nATOM 467 HD3 LYS A 26 11.979 -1.397 -5.235 1.00 0.00 H \nATOM 468 HE2 LYS A 26 10.776 -0.974 -7.390 1.00 0.00 H \nATOM 469 HE3 LYS A 26 10.239 0.588 -6.740 1.00 0.00 H \nATOM 470 HZ1 LYS A 26 13.043 -0.332 -7.088 1.00 0.00 H \nATOM 471 HZ2 LYS A 26 12.603 1.083 -6.255 1.00 0.00 H \nATOM 472 HZ3 LYS A 26 12.279 0.936 -7.916 1.00 0.00 H \nATOM 473 N GLY A 27 8.860 -5.990 -5.635 1.00 0.00 N \nATOM 474 CA GLY A 27 8.835 -7.203 -6.505 1.00 0.00 C \nATOM 475 C GLY A 27 10.264 -7.573 -6.904 1.00 0.00 C \nATOM 476 O GLY A 27 10.838 -6.984 -7.802 1.00 0.00 O \nATOM 477 H GLY A 27 9.080 -6.076 -4.688 1.00 0.00 H \nATOM 478 HA2 GLY A 27 8.386 -8.025 -5.966 1.00 0.00 H \nATOM 479 HA3 GLY A 27 8.259 -6.996 -7.394 1.00 0.00 H \nATOM 480 N ARG A 28 10.840 -8.543 -6.242 1.00 0.00 N \nATOM 481 CA ARG A 28 12.235 -8.961 -6.575 1.00 0.00 C \nATOM 482 C ARG A 28 12.327 -10.487 -6.649 1.00 0.00 C \nATOM 483 O ARG A 28 12.544 -10.995 -7.736 1.00 0.00 O \nATOM 484 CB ARG A 28 13.092 -8.426 -5.427 1.00 0.00 C \nATOM 485 CG ARG A 28 13.395 -6.945 -5.660 1.00 0.00 C \nATOM 486 CD ARG A 28 13.831 -6.298 -4.343 1.00 0.00 C \nATOM 487 NE ARG A 28 14.930 -5.364 -4.714 1.00 0.00 N \nATOM 488 CZ ARG A 28 15.256 -4.388 -3.912 1.00 0.00 C \nATOM 489 NH1 ARG A 28 16.102 -4.596 -2.941 1.00 0.00 N \nATOM 490 NH2 ARG A 28 14.735 -3.203 -4.079 1.00 0.00 N \nATOM 491 OXT ARG A 28 12.177 -11.121 -5.618 1.00 0.00 O \nATOM 492 H ARG A 28 10.354 -8.999 -5.523 1.00 0.00 H \nATOM 493 HA ARG A 28 12.549 -8.518 -7.506 1.00 0.00 H \nATOM 494 HB2 ARG A 28 12.558 -8.543 -4.495 1.00 0.00 H \nATOM 495 HB3 ARG A 28 14.019 -8.978 -5.383 1.00 0.00 H \nATOM 496 HG2 ARG A 28 14.188 -6.850 -6.389 1.00 0.00 H \nATOM 497 HG3 ARG A 28 12.509 -6.450 -6.026 1.00 0.00 H \nATOM 498 HD2 ARG A 28 13.006 -5.756 -3.901 1.00 0.00 H \nATOM 499 HD3 ARG A 28 14.199 -7.048 -3.659 1.00 0.00 H \nATOM 500 HE ARG A 28 15.410 -5.482 -5.561 1.00 0.00 H \nATOM 501 HH11 ARG A 28 16.502 -5.505 -2.811 1.00 0.00 H \nATOM 502 HH12 ARG A 28 16.352 -3.848 -2.326 1.00 0.00 H \nATOM 503 HH21 ARG A 28 14.087 -3.043 -4.824 1.00 0.00 H \nATOM 504 HH22 ARG A 28 14.984 -2.456 -3.464 1.00 0.00 H \nTER 505 ARG A 28 \nENDMDL \n';Mol1.loadMolecule(ChemDoodle.readPDB(Fmol, 1));Mol1.startAnimation();Mol1.stopAnimation();var resType = 'amino';function setDisp1(obj){Mol1.specs["proteins_display"+obj.value]=obj.checked;Mol1.repaint()}function setProj1(yesPers){Mol1.specs.projectionPerspective_3D=yesPers;Mol1.setupScene();Mol1.repaint()}function setColor1(obj){Mol1.specs.proteins_residueColor=resType=obj.value;Mol1.repaint()}function resColor1(obj){this.checked ? Mol1.specs.proteins_residueColor=resType : Mol1.specs.proteins_residueColor='none';Mol1.repaint()}</script><br><span class="meta">视图: <input type="radio" name="group2" onclick="setProj1(true)">投影 <input type="radio" name="group2" onclick="setProj1(false)" checked="">正交<br>着色: <input type="checkbox" onclick="Mol1.specs.macro_colorByChain=this.checked;Mol1.repaint()">按链 <input type="checkbox" onclick="resColor1(this)">按残基<br>模式: <input type="checkbox" value="Ribbon" checked="" onclick="setDisp1(this)">飘带 <input type="checkbox" value="Backbone" onclick="setDisp1(this)">骨架 <input type="checkbox" value="PipePlank" onclick="setDisp1(this)">管板 <input type="checkbox" checked="" onclick="Mol1.specs.proteins_ribbonCartoonize=this.checked;Mol1.repaint()">卡通<br>显示: <input type="checkbox" onclick="Mol1.specs.macro_showWater=this.checked;Mol1.repaint()">水分子 <input type="checkbox" onclick="Mol1.specs.atoms_nonBondedAsStars_3D=this.checked;Mol1.repaint()">非键原子 <input type="checkbox" onclick="Mol1.specs.atoms_displayLabels_3D=this.checked;Mol1.repaint()">名称<br>颜色: <input type="radio" value="amino" name="radio2" onclick="setColor1(this)">氨基酸 <input type="radio" value="shapely" name="radio2" onclick="setColor1(this)">形状 <input type="radio" value="polarity" name="radio2" onclick="setColor1(this)">极性 <input type="radio" value="acidity" name="radio2" onclick="setColor1(this)">酸性 <input type="radio" value="rainbow" name="radio2" onclick="setColor1(this)">彩虹<br>左键: 转动 滚轮: 缩放 双击: 自动旋转开关 Alt+左键: 移动</span><br><figurecaption>Fig.1</figurecaption></figure>作为示例, 我们只跑真空中的模拟, 100万步, 每步都保存到xtc文件. 这里要注意的是, 计算自由能时要尽量多保存轨迹, 越多越好, 因为构象越多, 得到的自由能形貌图越光滑.
计算相关性质
得到轨迹之后, 我们来计算回旋半径和RMSD(根据自己的需要选择分组):
<div class="highlight"><pre style="line-height:125%"><span style="color:#A2F">gmx</span> gyrate <span style="color:#666">-f</span> traj_comp.xtc
<span style="color:#A2F">gmx</span> rms <span style="color:#666">-f</span> traj_comp.xtc
</pre></div>这样就得到了gyrate.xvg, rmsd.xvg, 它们包含了每一步的总回旋半径Rg和RMSD值.
准备计算自由能的输入文件
要使用gmx sham计算自由能与Rg和RMSD的关系, 我们需要为它准备一个输入文件, 文件名称设为Rg-RMSD.xvg, 其中列出每一步对应的时间, Rg, RMSD值. 准备这种文件的简单方法是使用列编辑功能, 直接切出几列, 放到同一个文件中即可. 但如果每列的数据行数过多的话(在我们的例子中有100万行), 许多文本编辑器的列编辑功能可能会崩溃. 所以, 更稳定的方法是使用bash的paste命令, 它可以将两个文件按行合并在一起. 注意使用这个命令时, 文件的换行符应该是Linux下的换行符.
先将gyrate.xvg和rmsd.xvg文件中前面的注释和设置部分删掉, 只留下数据部分, 然后执行下面的命令
<div class="highlight"><pre style="line-height:125%"><span style="color:#A2F">paste</span> gyrate.xvg rmsd.xvg > Rg-RMSD.xvg
</pre></div>这样就得到了合并好的文件. 不过, 文件中还含有我们不需要的数据, X/Y/Z三个方向的Rg值和RMSD对应的时间值, 将这些列直接使用列编辑功能删除就好了.
有了输入文件后, 就可以计算自由能了:
<div class="highlight"><pre style="line-height:125%"> <span style="color:#A2F">gmx</span> sham <span style="color:#666">-f</span> Rg-RMSD.xvg <span style="color:#666">-ls</span> <span style="color:#666">-b</span> 20 <span style="color:#666">-ngrid</span> 200 200 200 <span style="color:#666">-nlevels</span> 100
</pre></div>这里指定从20 ps开始计算, 是因为从RMSD曲线很容易看出, 20 ps后RMSD才稳定. 如果不忽略前面未平滑部分, 计算出的自由能范围可能过大, 对后面的作图不利. 后面的选项根据你自己的测试来设置就好.
作图
得到的gibbs.xpm文件就包含了自由能与Rg和RMSD的关系, 用它就可以作图了. 如果是作二维图的话, gmx xpm2ps就可以了. 要作三维填色图的话, 麻烦点, 需要使用Origin, MatLab之类的作图软件. 但这些软件在作图的时候无法将自由能形貌图和蛋白一起渲染. 要达到这个目的, 我们可以使用PyMOL.
我以前曾经示例过如何利用PyMOL制作反应势能面示意动画, 将那里的脚本稍加修改就能满足我们的需要. 不过在作图之前, 我们需要将xpm中的数据提取出来. 这可以使用前面提到的那篇教程中提供的一个脚本, 但那个脚本只适用于水平数低于80的情况. 我们这里使用的水平数为100, 所以需要对脚本稍加修改. 此外, 如果得到的图形尖峰太多, 可以对数据进行简单的平滑, 这样图形看起来更漂亮.
然后再载入蛋白, 调整好位置, 将其与形貌图放在一起渲染.
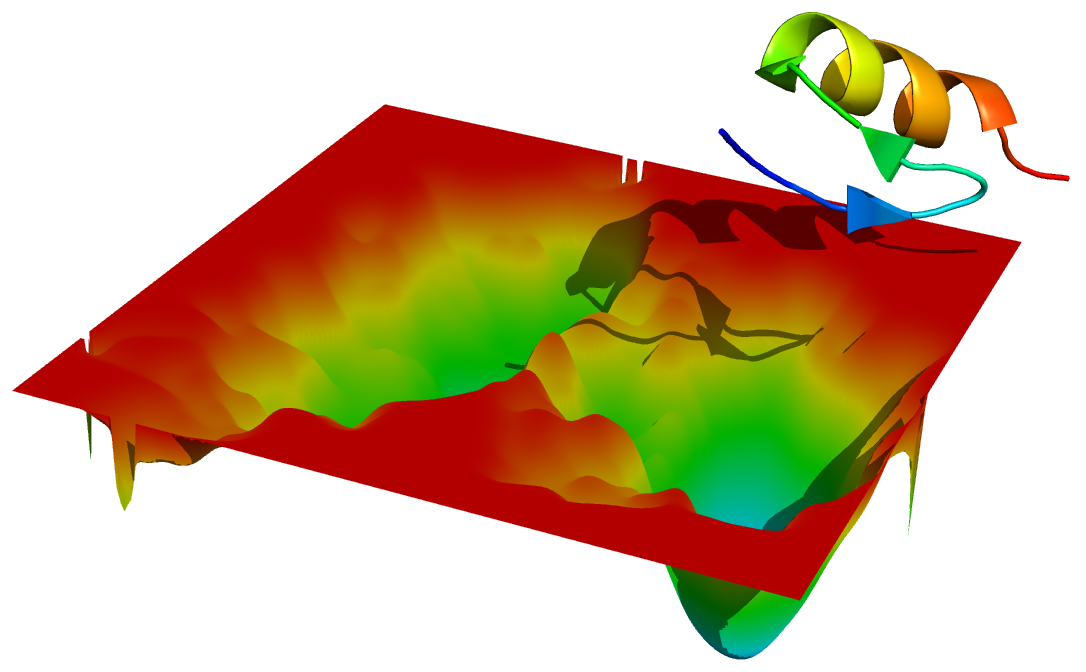
看, 蛋白在形貌图上有投影的.
当然, 你还可以将其他有代表性的构象放在自由能形貌图的不同位置, 这样信息量就更大了. 其他的很多设置都可以试着调整. 这些就不赘述了.
不过这种作图方法的一个缺点就是不好加坐标轴, 需要额外的代码才可以.
代码(略)

